A team from Columbia University present a microfluidic device that can capture single-cell lysates in picotire plates and produce single-cell 3' tag RNA-Seq library prep at $0.10 cents per cell: Scalable microfluidics for single cell RNA printing and sequencing. They discuss two methods - RNA “printing” on glass and RNA capture on beads, but the paper focuses on the second of these. This is an early paper with two experiments each of which could be
improved by another round or two in the lab. However in this fast moving
field I can’t help but wonder if this is a method that will never be
commercialised and if the authors simply want to make their mark? I think the fact that single-cell methods are appearing thick and fast shows how much this space there still is to innovate into. And for the investment pundits looking to Fluidigm et al, there is a danger that a clever molecular biology approach could allow single cell without any hardware - imagine library prep inside a cell?
How does it work: In the RNA capture on beads method cells are loaded into microwells, then barcoded oligo-dT coated beads are loaded such that one fills each picotitre well. cDNA synthesis on these beads generates a barcoded cDNA ready for downstream RNA-seq library prep. In the paper they provide proof-of-concept data on a five lane chip with >10,000 microwells, cells are loaded at low concentration and Poisson statistics ensure that >95% of wells with have 1 or 0 cells. Wells are sized such that only a single bead can also be accommodated. The chip is hydrated and washed with PBS, cells were loaded and settle by gravity, excess cells are removed by further PBS washes. The microwells are “closed” by oil and freeze-thaw was used to lyse cells releasing mRNA onto the beads. After mRNA capture all subsequent steps are performed with the chip “open”. They used the Cel-seq protocol where 1st and 2nd strand cDNA synthesis are followed by IVT. The resulting aRNA is collected and is used for a second RT using random hexamers, and this is followed by PCR to produce barcoded RNA-seq libraries for sequencing. Phew!
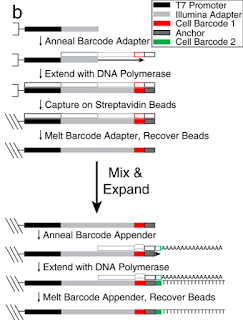
Barcode production: They employ a very clever combinatorial barcoding method that allows just 106 oligos to produce 960 barcodes. First, beads are coated with oligos containing a T7 promoter and Illumina adaptor. Second, the beads are plated to allow 96 individual barcode adaptors to be annealed to these covalently attached oligos, and primer extension creates the full-length covalently attached 1st barcode oligo. Third, beads are pooled for a final step similar to round two but this a second round of primer extension using one of ten secondary barcoding oligo creates a final pool of 960 uniquely barcoded beads. Phew again!!
In the paper they describe an experiment using U87 (Glioma) and MCF10a (Breast cancer) cells and comparison to bulk U87 RNA-seq and a GEO MCF7 dataset. Up to 50% of single-cells produce only low-coverage data, suggesting there is significant room for improvement. Cross-talk of some kind did occur with beads that did not appear to be co-located with a cell (as assessed by imaging) but that generated reads; however at 200-300x lower coverage. Although the method is a 3’ tag counting one only 500-600 genes were detected in each cell, perhaps another area with room for improvement?
This could well be another method to watch. The manufacutre of thier barcodes is a nice piece of work and could easily be applied to single-cells in microtitre plates using flow-cytometry, making it amenable to pretty much any lab.
No comments:
Post a Comment
Note: only a member of this blog may post a comment.